$ rails console >> 1.year.from_now => Sun, 13 Mar 2011 03:38:55 UTC +00:00 >> 10.weeks.ago => Sat, 02 Jan 2010 03:39:14 UTC +00:00 >> 1.hour.from_now => Sun, 12 Aug 2012 09:52:39 UTC +00:00 >> 30.seconds.from_now => Sun, 12 Aug 2012 08:54:00 UTC +00:00
my personal notepad - for all the things I used to write on pieces of paper ( which where never seen again esp. when I needed them ) ...
Sunday, August 12, 2012
ruby rails - examples for date time helpers
with the time helpers it is amazingly easy to deal with dates and times (add, diff...)
Thursday, August 9, 2012
rails haml - translations
<%= yield %> --> .content#content= yield
<%= csrf_meta_tag %> --> = csrf_meta_tag
<%= csrf_meta_tag %> --> = csrf_meta_tag
use stylesheets
<%= stylesheet_link_tag 'blueprint/screen', :media => 'screen' %> --> = stylesheet_link_tag 'main'
ruby rails - set up autotest
http://www.andrewsturges.com/2011/06/installing-autotest-with-rails-31-and.html
+ run: rails g rspec:install (rspec-rails have to be installed)
+ run: rails g rspec:install (rspec-rails have to be installed)
Wednesday, August 8, 2012
ruby - problems with rvm
if rvm does not set the right ruby version:
http://stackoverflow.com/questions/9056008/installed-ruby-1-9-3-with-rvm-but-command-line-doesnt-show-ruby-v/9056395#9056395
http://stackoverflow.com/questions/9056008/installed-ruby-1-9-3-with-rvm-but-command-line-doesnt-show-ruby-v/9056395#9056395
Saturday, August 4, 2012
R ggplot - rebuild cdc obesity maps - 1984-2011
redoing the cdc obesity maps with ggplot2
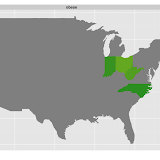 |
| rebuild cdc obesity maps with ggplot |
Slideshow
- first obese rates
- second overweight rates
- and if the slide show does not work - here is the link to the
pictures
Table of Contents
1 get the data
- I downloaded the data from http://www.cdc.gov/brfss/technical_infodata/surveydata.htm
- for the years 1984 - 1997 I use read.xport() (foreign package) on the sas xpt files
- then the data sets became to large, so I used the ascii files read.fortran() and choose just a few columns
- here is a resulting example data set (2006 - I computed the bmi2 column for checking)
- 2012-09: I added the maps for 2011 since the new data were out
head(x2006)
State month day year age weight height sex htm wkg bmi bmigr bmirisk 1 1 5 2 2006 66 263 503 2 160 11955 4669 3 2 2 1 9 19 2006 56 290 603 1 191 13182 3632 3 2 3 1 12 12 2006 40 230 511 1 180 10455 3215 3 2 4 1 4 29 2006 38 320 603 1 191 14545 4008 3 2 5 1 4 29 2006 52 120 504 2 163 5455 2064 1 1 6 1 8 2 2006 32 165 510 2 178 7500 2372 1 1 heightcm weightkg bmi2 1 160.02 119.29417 46.58764 2 190.50 131.54110 36.24695 3 180.34 104.32570 32.07799 4 190.50 145.14880 39.99664 5 162.56 54.43080 20.59763 6 177.80 74.84235 23.67467
2 compute rates and plot the graphs
library(ggplot2)
library(scales)
library(plyr)
library(maps)
## map of the states (part of the map package)
states_map <- map_data("state")
states_map$region <- factor(states_map$region)
## got fips form here and saved it as txt; http://www.epa.gov/enviro/html/codes/state.html
fips <- read.table("states.txt",sep="\t",header=T)
fips$State.Name <- tolower(as.character(fips$State.Name))
## build the graphs
filenames <- paste("bmi",1984:2010,".rdata",sep="")
for(file in filenames){
load(file)
year <- substr(file,4,7)
x <- get(paste("x",year,sep=""))
## for adding the year to the plot
testdf <- data.frame(x2=-70,y2=49,year=year)
## for the first 4 years was no bmi in the data set
## I named my computed one "bmi" so I need another "bmi2" for the loop, not very sophisticated,
## but it works
if(!("bmi2" %in% names(x))){
print(file)
x$bmi2 <- x$bmi
}
## bmi groups
x$bmi2gr <- cut(x$bmi2,breaks=c(0,25,30,300),include.lowest=T,labels=c("1","2","3"))
## count
x <- ddply(x,.(State),transform,perstate=sum(!is.na(bmi2)))
x <- ddply(x,.(State,bmi2gr),transform,perstate.gr=sum(!is.na(bmi2)))
dats <- unique(x[,c("State","bmi2gr","perstate","perstate.gr")])
dats <- dats[!is.na(dats$bmi2gr),]
## percents
dats$perc <- dats$perstate.gr/dats$perstate
dats$ow <- as.numeric(dats$bmi2gr) > 1
## I just want the obese and overweight
## >= 25
dats2 <- dats[dats$ow==T,]
dats2 <- ddply(dats2,.(State),summarize,perc=sum(perc))
dats2$gr <- "ow"
## >= 30
dats3 <- dats[dats$bmi2gr=="3",c("State","perc")]
dats3$gr <- "obese"
dats <- rbind(dats2,dats3)
## identify the states in the data set using the region names in the map (fips coded)
dats <- merge(dats,fips[,2:3],by.x="State",by.y="FIPS.Code",all=T)
dats$gr[is.na(dats$gr)] <- "obese"
dats$State.Name <- factor(dats$State.Name)
## graph
ggplot(dats[dats$gr=="obese",],aes(map_id = State.Name)) +
geom_map(aes(fill=perc),colour="black",map = states_map) +
expand_limits(x = states_map$long, y = states_map$lat) +
scale_fill_gradientn(limits=c(0.1,0.7),colours=cols,guide = guide_colorbar(),na.value="grey50") +
geom_text(data=testdf,aes(x=x2,y=y2,label=year),inherit.aes=F)
## save image
ggsave(file=paste("obese",substr(file,4,7),".png",sep=""))
}
output
[1] "bmi1984.rdata" Saving 12.7 x 7.01 in image [1] "bmi1985.rdata" Saving 12.7 x 7.01 in image [1] "bmi1986.rdata" Saving 12.7 x 7.01 in image [1] "bmi1987.rdata" Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image Saving 12.7 x 7.01 in image
Date: 2012-08-04 21:24:07 CEST
Author: mandy
Org version 7.8.02 with Emacs version 23
Validate XHTML 1.0
Labels:
cdc
,
obesity
,
R
,
R ggplot
,
R graphics
,
R map graphics
,
R maps
Subscribe to:
Comments
(
Atom
)
